# Load packages
library(sf)
library(terra)
library(lidR)
library(lidRmetrics)Excercise Solutions
Resources
1-LAS
E1.
Using the plot() function plot the point cloud with a different attribute that has not been done yet
Try adding axis = TRUE, legend = TRUE to your plot argument plot(las, axis = TRUE, legend = TRUE)
Code
las <- readLAS(files = "data/fm_norm.laz")Code
plot(las, color = "ReturnNumber", axis = TRUE, legend = TRUE)E2.
Create a filtered las object of returns that have an Intensity greater that 50, and plot it.
Code
las <- readLAS(files = "data/fm_norm.laz")Code
i_50 <- filter_poi(las = las, Intensity > 50)
plot(i_50)E3.
Read in the las file with only xyz and intensity only. Hint go to the lidRbook section to find out how to do this.
Code
las <- readLAS(files = "data/fm_norm.laz", select = "xyzi")2-DTM
E1.
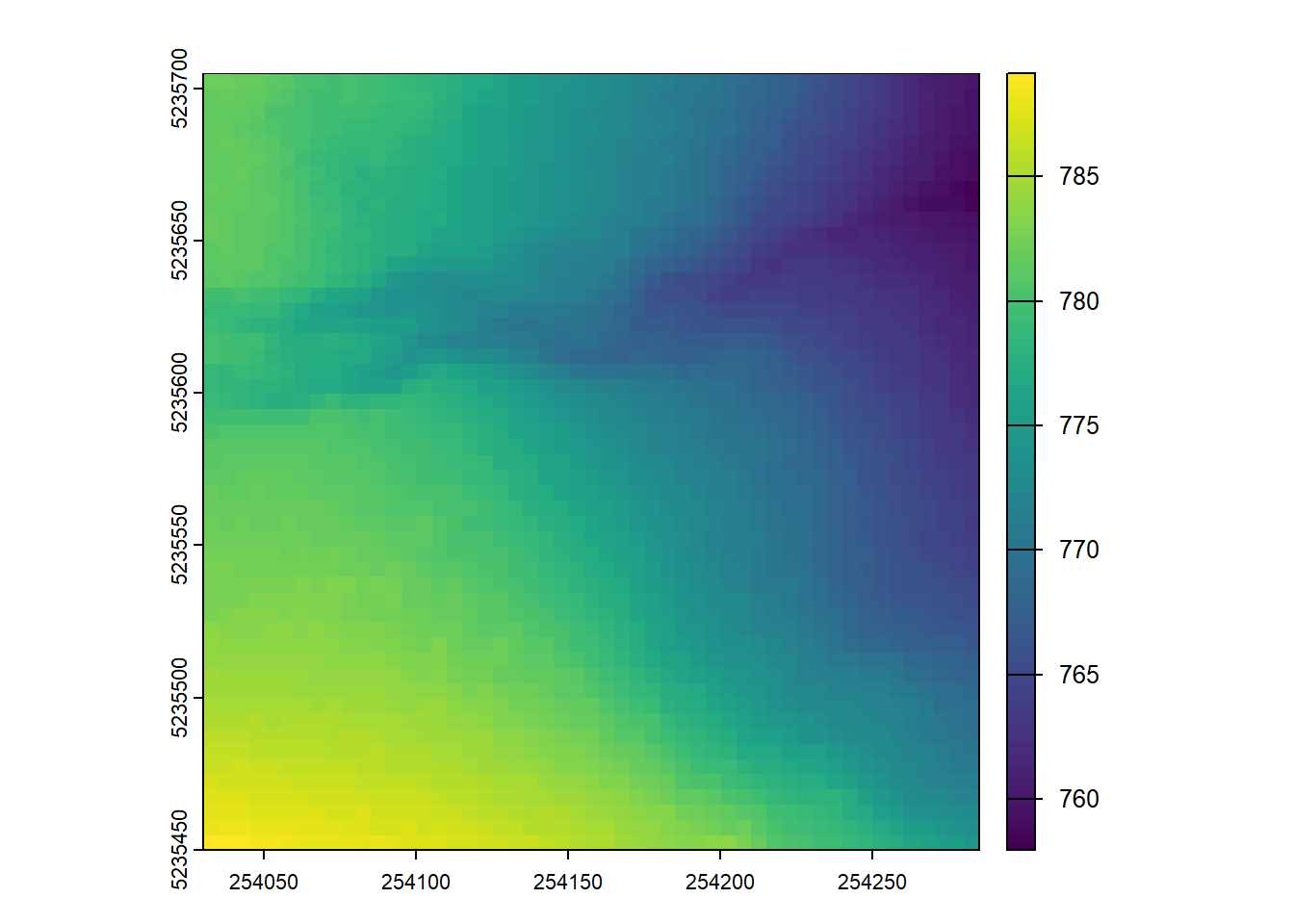
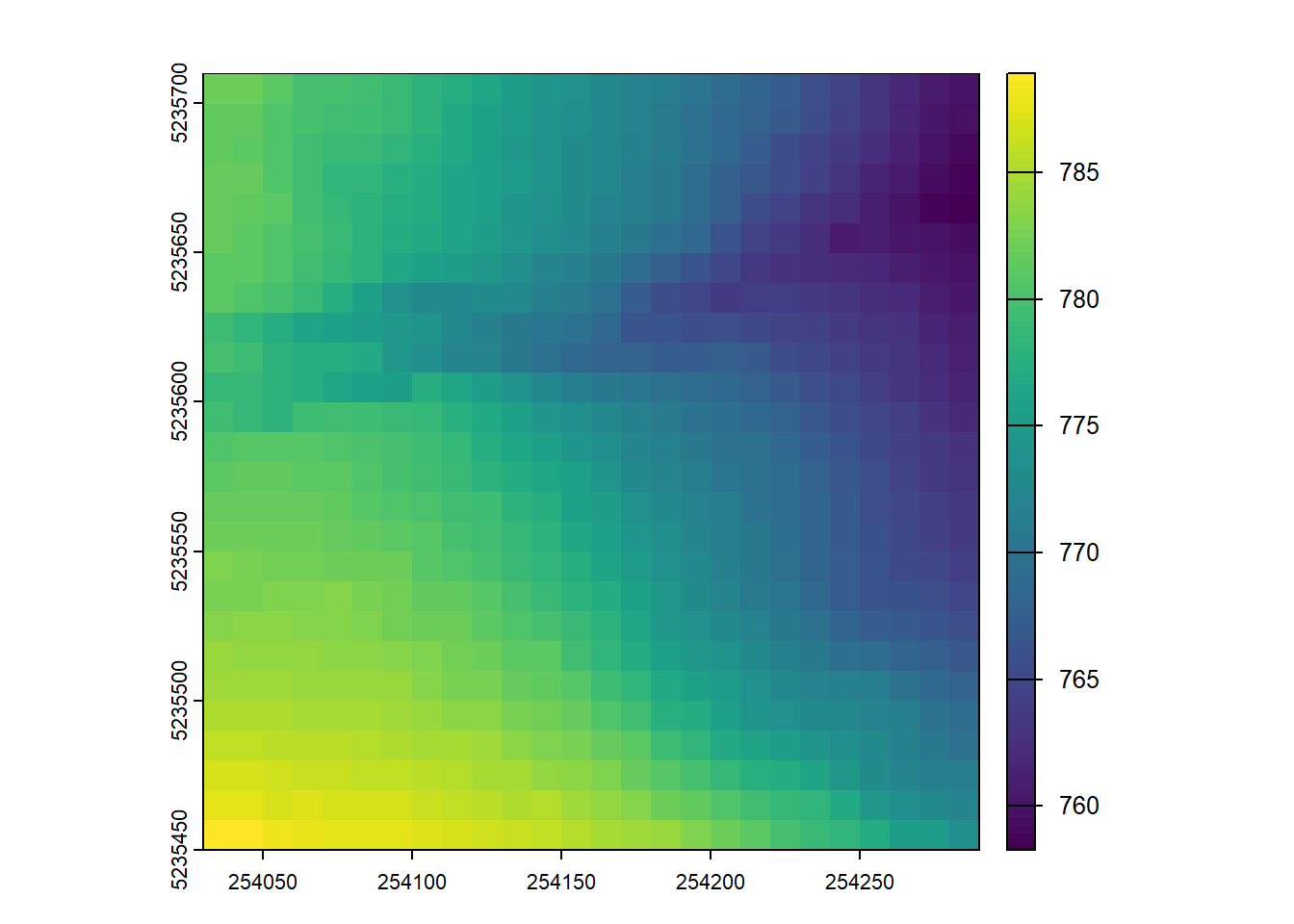
Compute two DTMs for this point cloud with differing spatial resolutions, plot both e.g. `plot(dtm1)
E2.
Now use the plot_dtm3d() function to visualize and move around your newly created DTMs
Code
plot_dtm3d(dtm_5)3-CHM
E1.
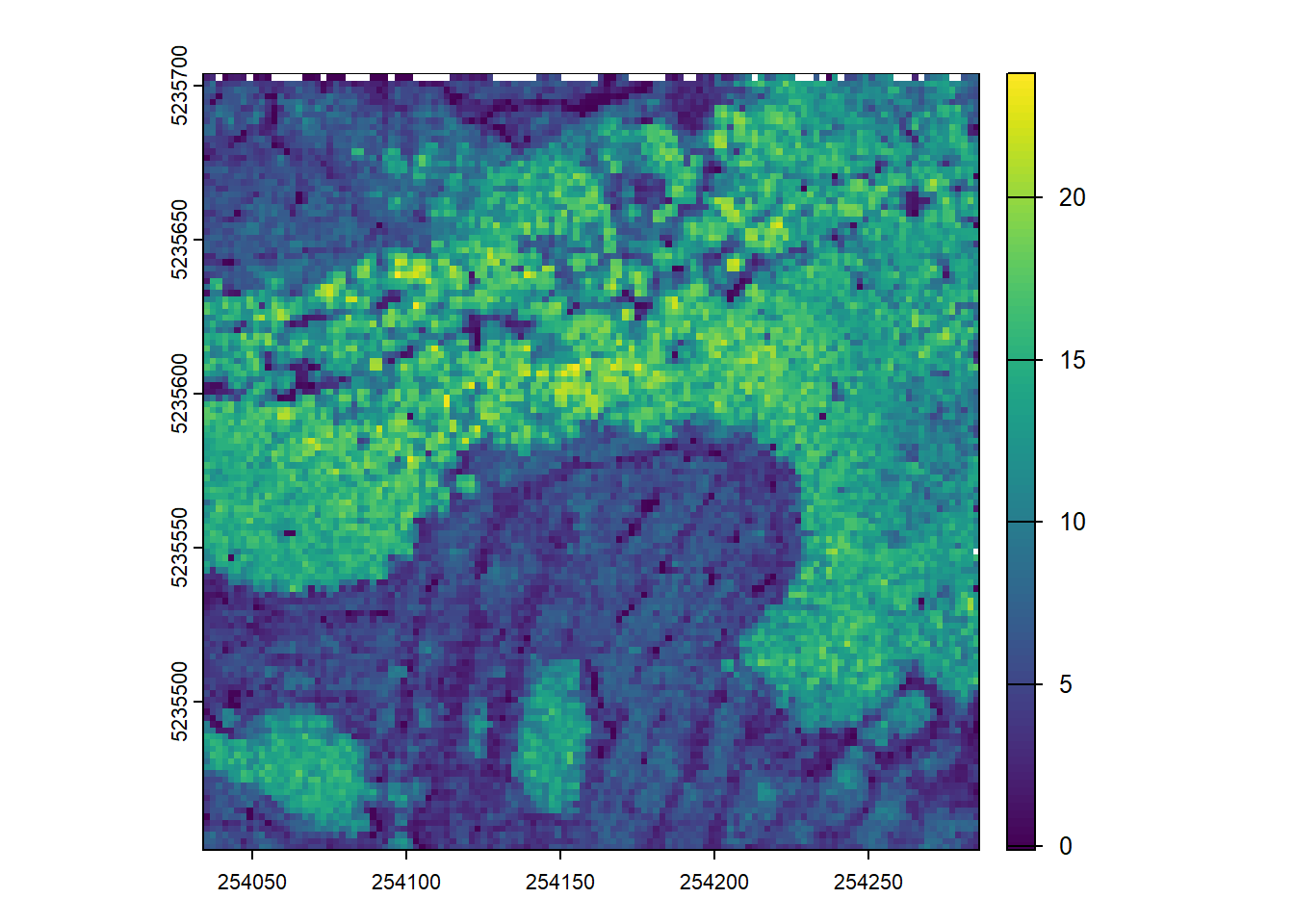
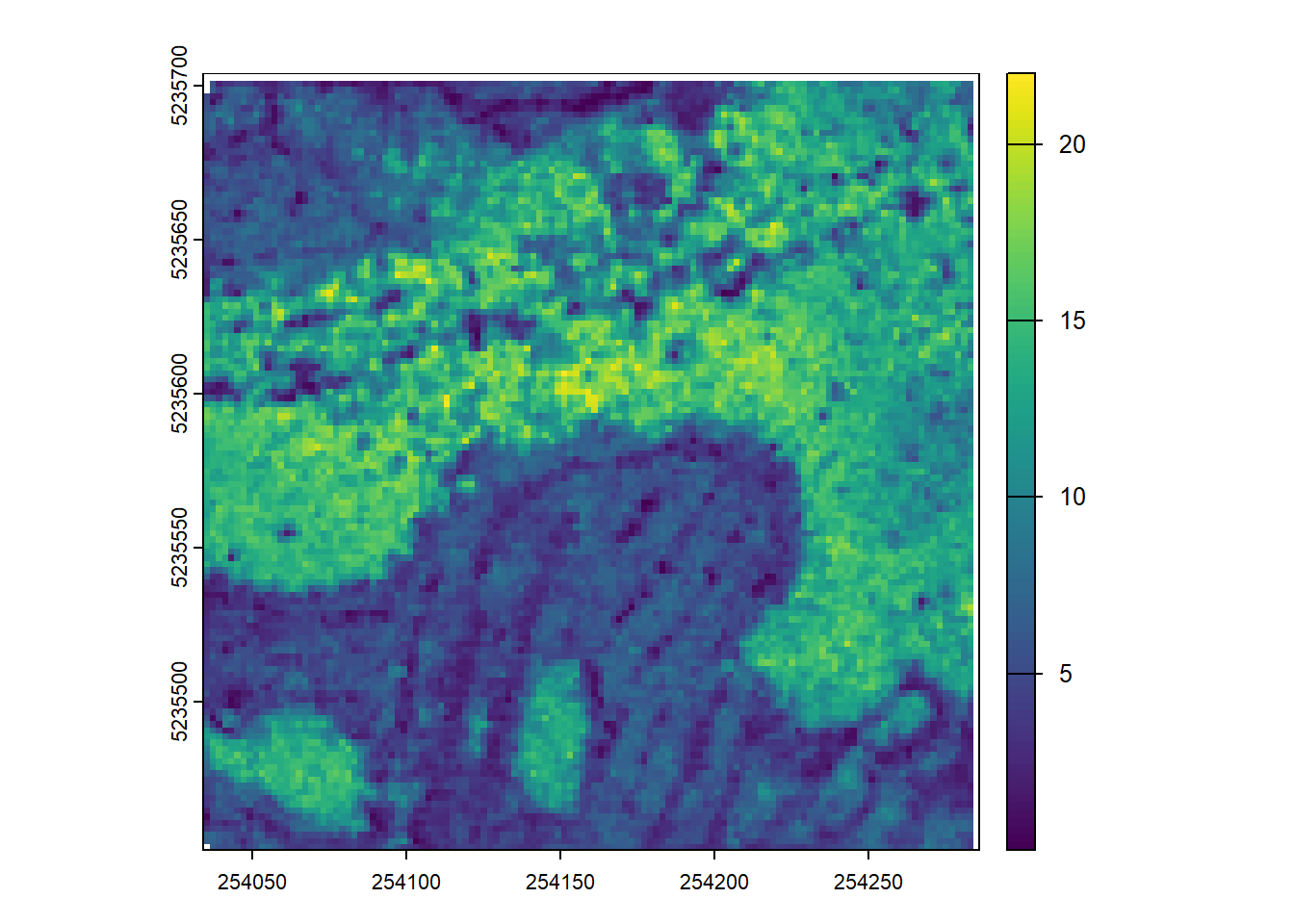
Using the p2r() and pitfree() (defining your own arguments) create two CHMs with the same resolution and plot them. What differences do you notice?
Code
las <- readLAS(files = "data/fm_norm.laz")Code
chm_p2r <- rasterize_canopy(las=las, res=2, algorithm=p2r())
thresholds <- c(0, 5, 10, 20, 25, 30)
max_edge <- c(0, 1.35)
chm_pitfree <- rasterize_canopy(las = las, res = 2, algorithm = pitfree(thresholds, max_edge))
plot(chm_p2r)Code
plot(chm_pitfree)E2.
Using terra::focal() with the w = matrix(1, 5, 5) and fun = max, plot a manipulated CHM using one of the CHMs you previously generated.
E3.
Create a 10 m CHM using a algorithm of your choice. Would this information still be useful at this scale?
4-METRICS
E1.
Generate another metric set provided by the lidRmetrics package (voxel metrics will take too long)
Code
las <- readLAS(files = "data/fm_norm.laz")Code
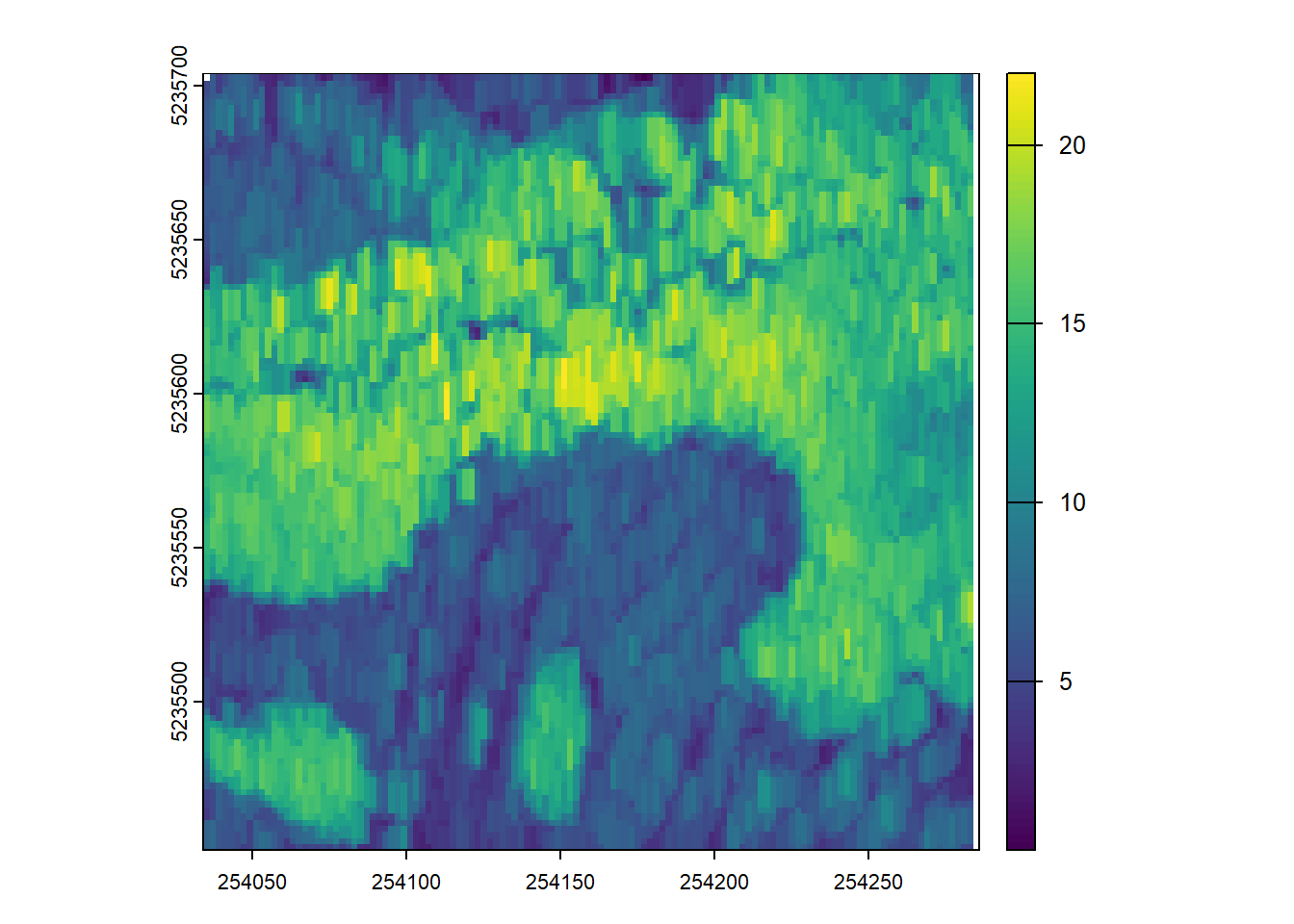
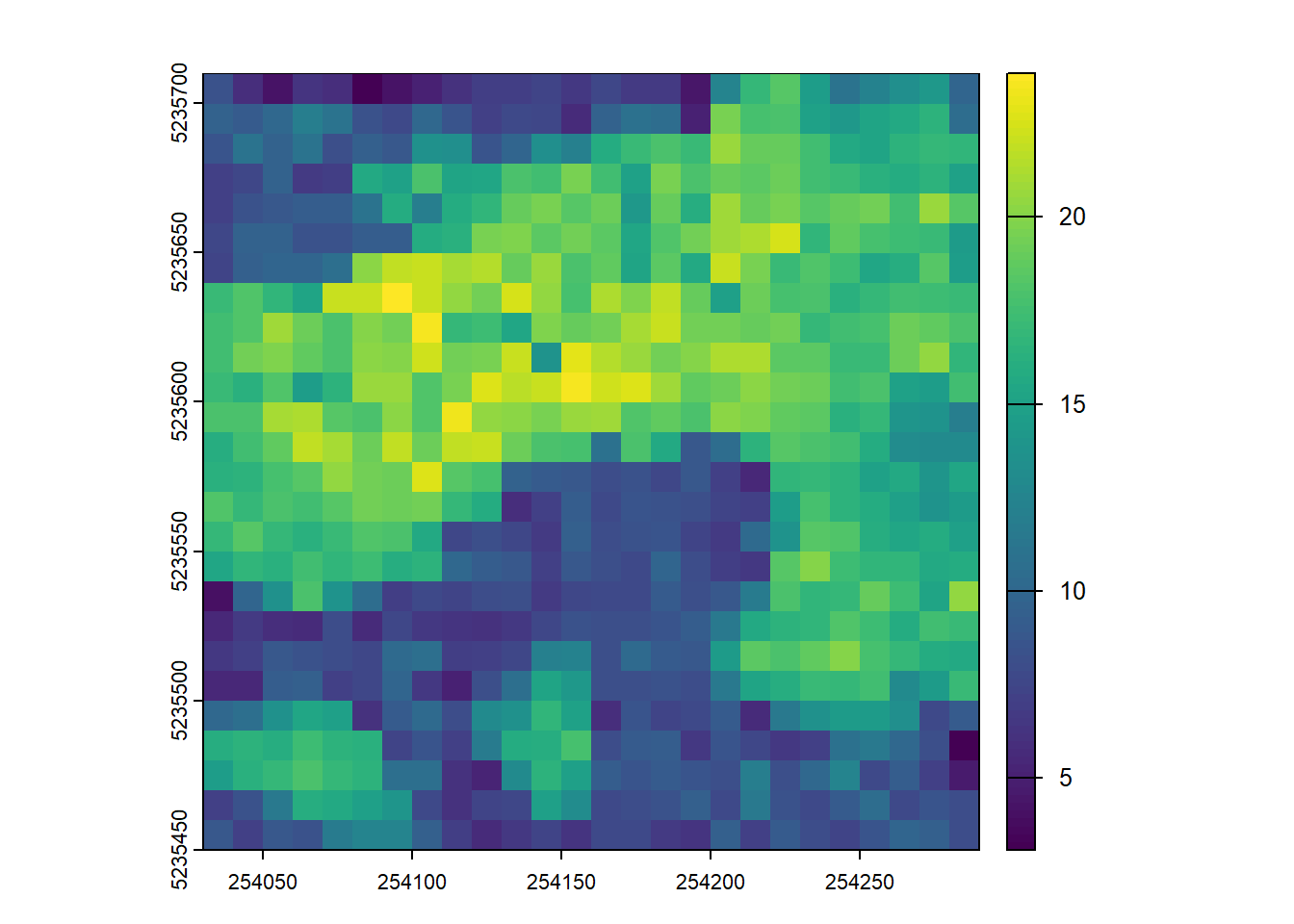
dispersion <- pixel_metrics(las, ~metrics_percentiles(z = Z), res = 20)E2.
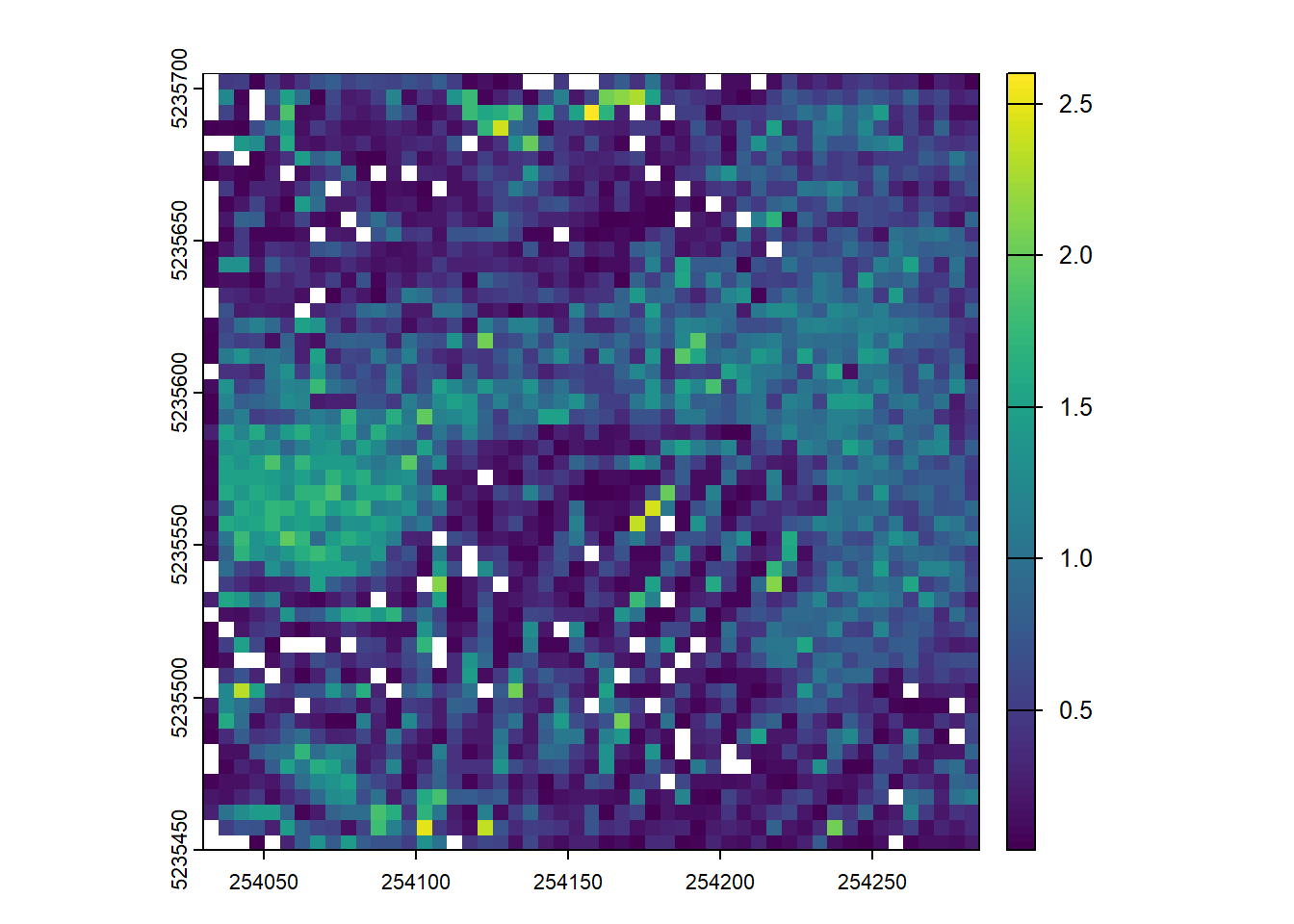
Map the density of ground returns at a 5 m resolution. Hint filter = -keep_class 2
E3.
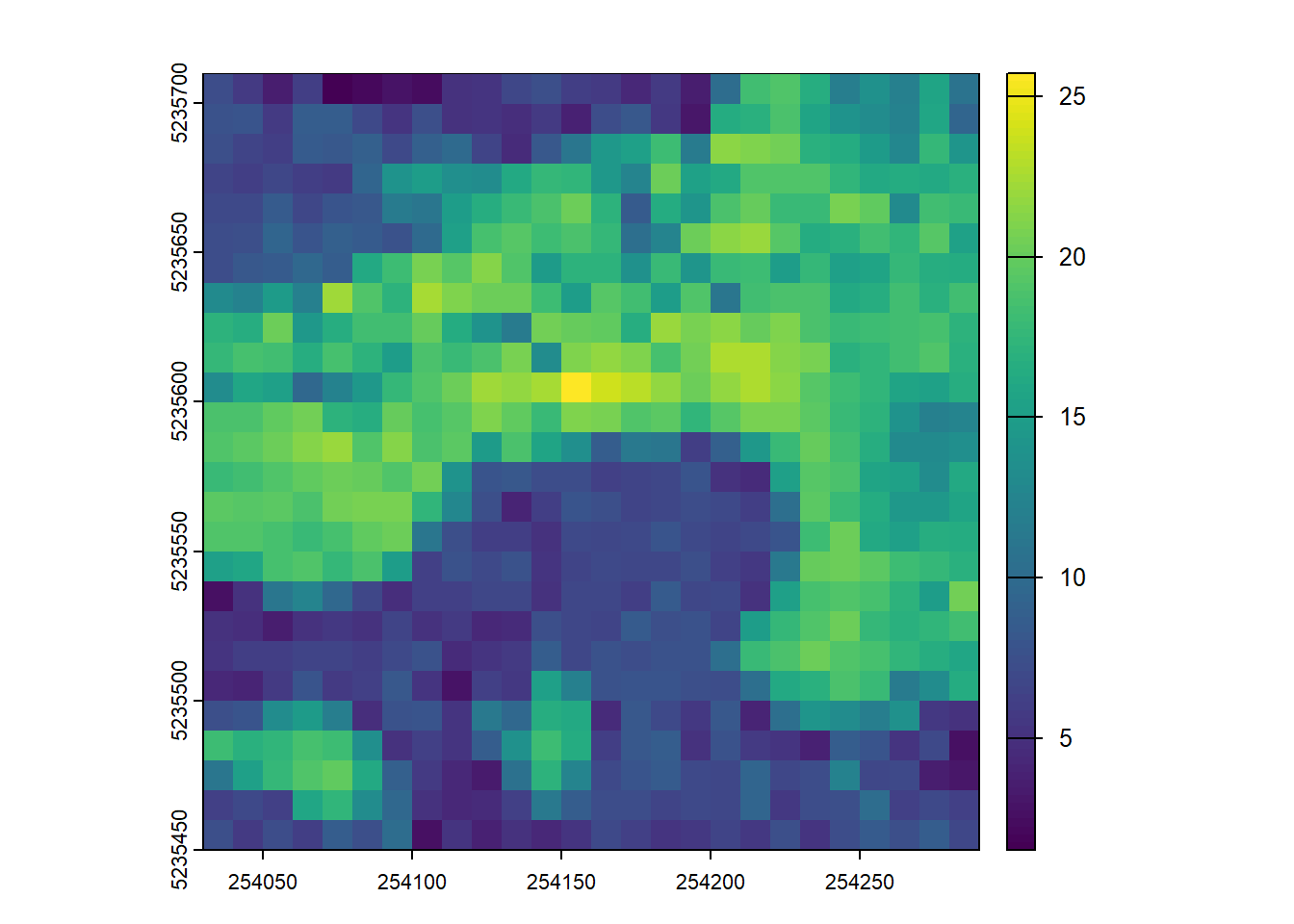
ssuming that biomass is estimated using the equation B = 0.5 * mean Z + 0.9 * 90th percentile of Z applied on first returns only, map the biomass.
6-LASCATALOG
E1.
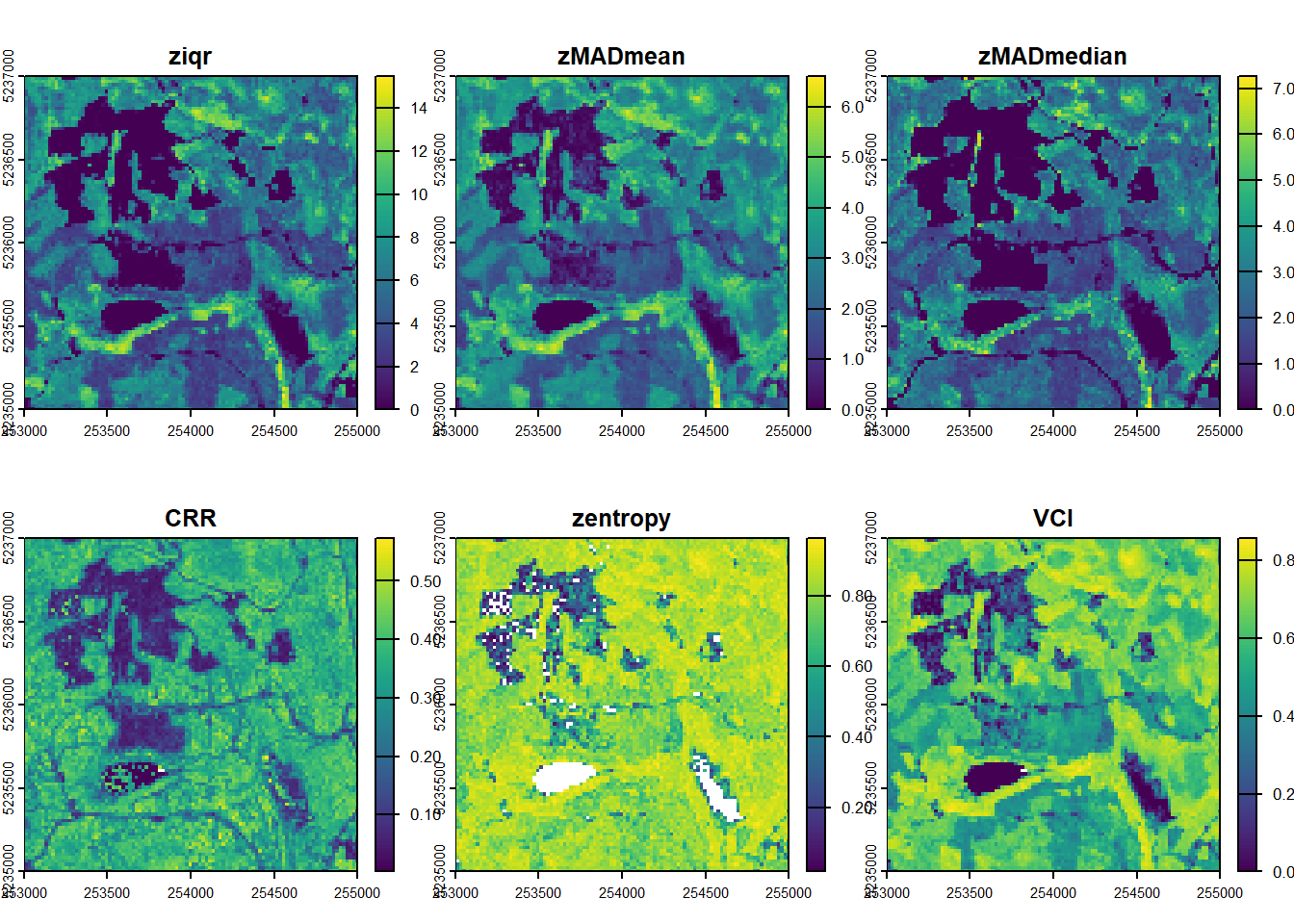
Calculate a set of metrics from the lidRmetrics package on the catalogue (voxel metrics will take too long)
Code
ctg <- readLAScatalog(folder = "data/ctg_norm")
dispersion <- pixel_metrics(ctg, ~metrics_dispersion(z = Z, dz = 2, zmax = 30), res = 20)#> Chunk 1 of 16 (6.2%): state ✓
#>
Chunk 2 of 16 (12.5%): state ✓
#>
Chunk 3 of 16 (18.8%): state ✓
#>
Chunk 4 of 16 (25%): state ✓
#>
Chunk 5 of 16 (31.2%): state ✓
#>
Chunk 6 of 16 (37.5%): state ✓
#>
Chunk 7 of 16 (43.8%): state ✓
#>
Chunk 8 of 16 (50%): state ✓
#>
Chunk 9 of 16 (56.2%): state ✓
#>
Chunk 10 of 16 (62.5%): state ✓
#>
Chunk 11 of 16 (68.8%): state ✓
#>
Chunk 12 of 16 (75%): state ✓
#>
Chunk 13 of 16 (81.2%): state ✓
#>
Chunk 14 of 16 (87.5%): state ✓
#>
Chunk 15 of 16 (93.8%): state ✓
#>
Chunk 16 of 16 (100%): state ✓
#>
plot(dispersion)E2.
Read in the non-normalized las catalog filtering the point cloud to only include first returns.
Code
ctg <- readLAScatalog(folder = "data/ctg_class")
opt_filter(ctg) <- "-keep_first -keep_class 2"E3.
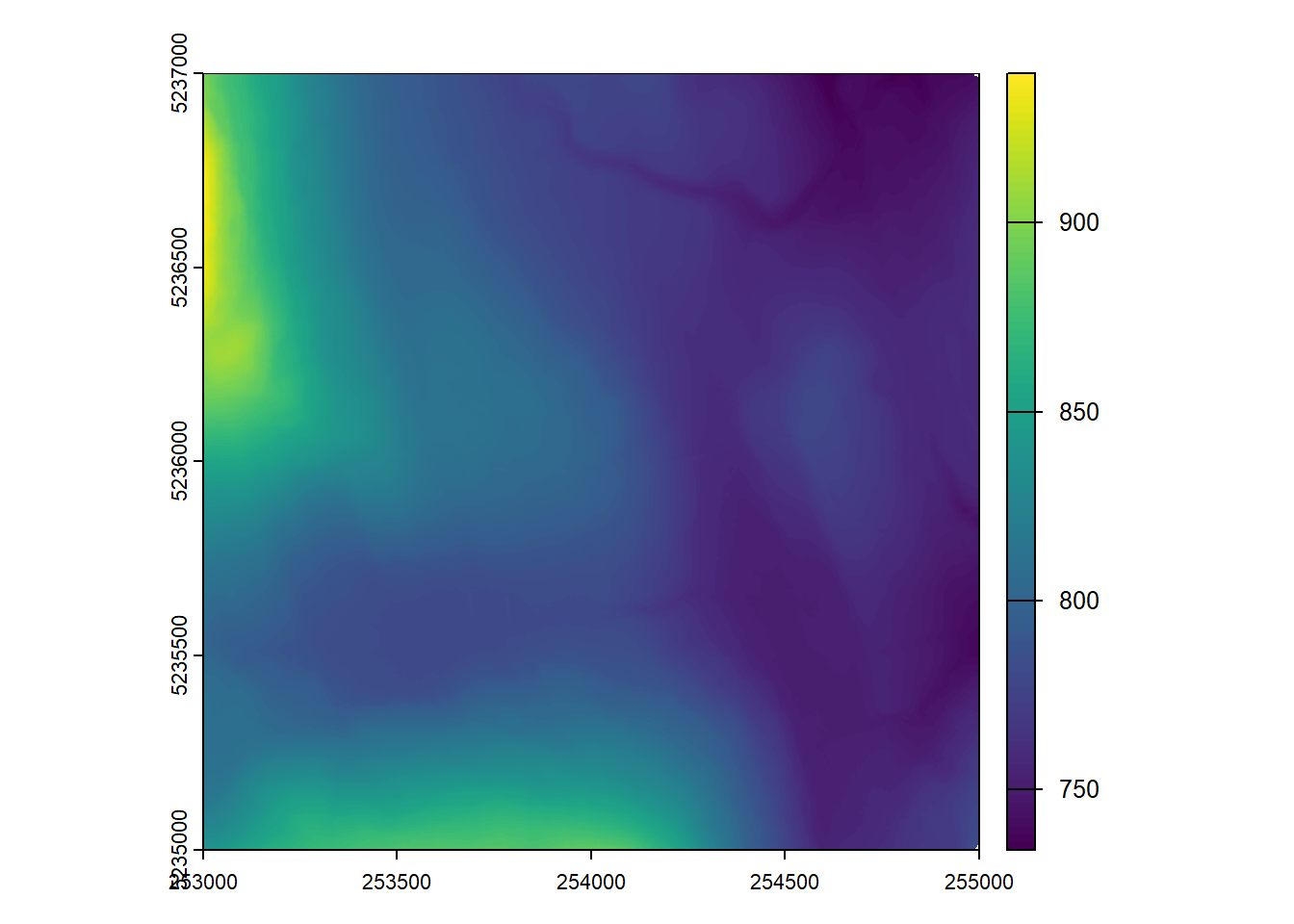
Generate a DTM at 1m spatial resolution for the provided catalog with only first returns.
Code
dtm_first <- rasterize_terrain(ctg, res = 1, algorithm = tin())#> Chunk 1 of 16 (6.2%): state ✓
#>
Chunk 2 of 16 (12.5%): state ✓
#>
Chunk 3 of 16 (18.8%): state ✓
#>
Chunk 4 of 16 (25%): state ✓
#>
Chunk 5 of 16 (31.2%): state ✓
#>
Chunk 6 of 16 (37.5%): state ✓
#>
Chunk 7 of 16 (43.8%): state ✓
#>
Chunk 8 of 16 (50%): state ✓
#>
Chunk 9 of 16 (56.2%): state ✓
#>
Chunk 10 of 16 (62.5%): state ✓
#>
Chunk 11 of 16 (68.8%): state ✓
#>
Chunk 12 of 16 (75%): state ✓
#>
Chunk 13 of 16 (81.2%): state ✓
#>
Chunk 14 of 16 (87.5%): state ✓
#>
Chunk 15 of 16 (93.8%): state ✓
#>
Chunk 16 of 16 (100%): state ✓
#>
plot(dtm_first)